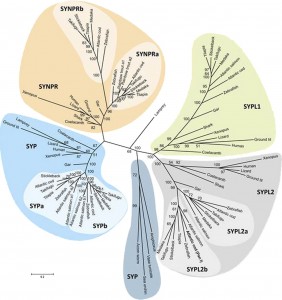
In this study, we aimed at gaining more knowledge of the enigmatic role of the synaptophysins in the adaptive evolution of stationary and migratory populations of Atlantic cod (Gadus morhua L.).
The synaptophysin protein family consists of membrane proteins involved in vesicle-trafficking events, but the physiological function of several members is unknown. Notably, a member of the synaptophysins, pantophysin (Pan I), has been shown to be highly polymorphic among populations of Atlantic cod and we have also previously revealed that the stationary and migratory ecotypes of Atlantic cod display near fixed alleles at this locus (e.g. Fevolden et al 2012). The divergence between the ecotypes of Atlantic cod at this locus has been suggested to reflect local adaptation, e.g. to alternative regimes of light, during settling of the YOY and to vertical behavior of the adults. However, whether the adaptive divergence between the ecotypes at the Pan I locus is caused by “divergence hitch-hiking” of Pan I with other genes/synaptophysins (e.g. rhodopsin, known to be directly linked to perception of alternative wavelengths of light) or by directional selection against unfavorable Pan I alleles during settling and vertical migration also remains to be illuminated. Additionally, in order to understand the adaptive significance of these genes, we also need a better understanding of the evolution of the gene family and whether the protein phenotypes produced by the allelic variants differ in composition.